Multiscale affinity maturation simulations to elicit broadly neutralizing antibodies against HIV
Simone Conti, Victor Ovchinnikov, Jonathan G. Faris, Arup K. Chakraborty, Martin Karplus, and Kayla G. Sprenger
PLOS Computational Biology, 2022, 18(4), pp e1009391.
DOI:10.1371/journal.pcbi.1009391 | Find on RG
Abstract:
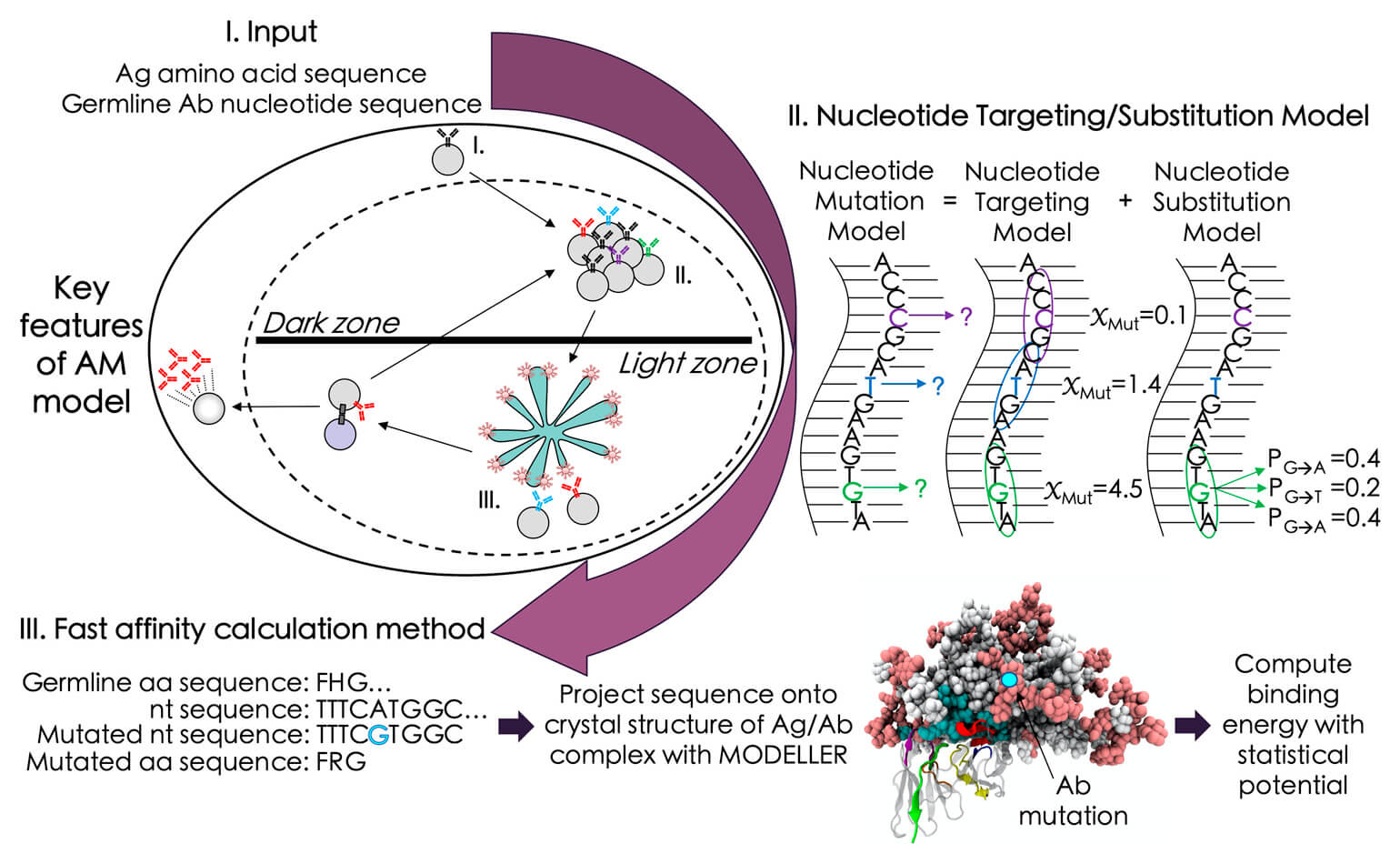
The design of vaccines against highly mutable pathogens, such as HIV and influenza, requires a detailed understanding of how the adaptive immune system responds to encoun- tering multiple variant antigens (Ags). Here, we describe a multiscale model of B cell recep- tor (BCR) affinity maturation that employs actual BCR nucleotide sequences and treats BCR/Ag interactions in atomistic detail. We apply the model to simulate the maturation of a broadly neutralizing Ab (bnAb) against HIV. Starting from a germline precursor sequence of the VRC01 anti-HIV Ab, we simulate BCR evolution in response to different vaccination pro- tocols and different Ags, which were previously designed by us. The simulation results pro- vide qualitative guidelines for future vaccine design and reveal unique insights into bnAb evolution against the CD4 binding site of HIV. Our model makes possible direct compari- sons of simulated BCR populations with results of deep sequencing data, which will be explored in future applications.
